Structural immunology
We investigate the structures of immune system components using a combination of cryo-electron microscopy (cryoEM), cryo-electron tomography (cryoET) and X-ray crystallography.
CryoEM is generally used on purified solution-phase biomolecules, such as the pentraxins C-reactive protein [25] and PTX3 [28], components of our innate immune system. These solution phase cryoEM structures usually represent the inactive state of immune components, which often require ligand binding to become activated.
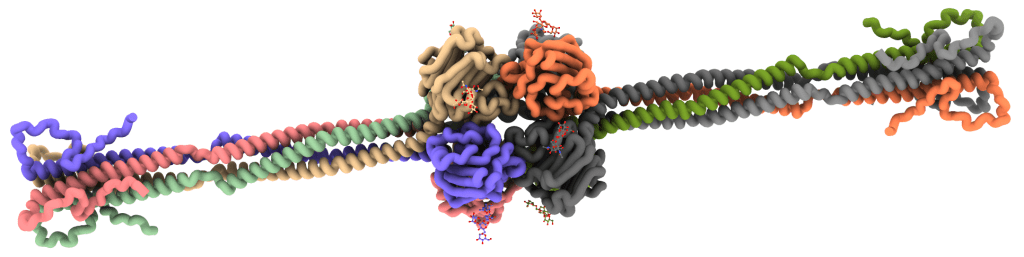
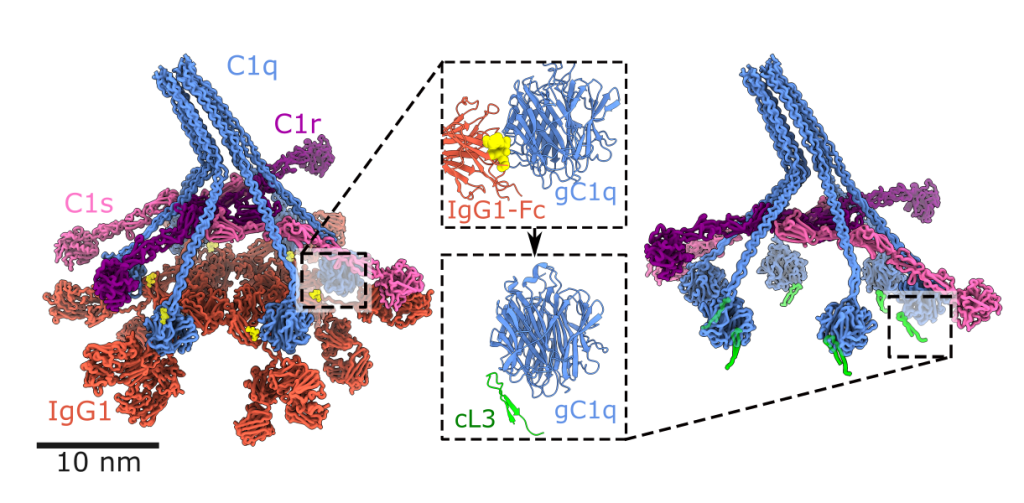
To investigate surface-associated complexes, we use cryoET, which provides contextual information and allows subtomogram averaging. CryoET is typically much lower resolution that cryoEM but the added contextual information can yield surprising unique insights into immune system activation by pentraxins [45], IgG1 [17], IgG3 [33] or IgM [21] antibodies, or other biomolecules.
X-ray crystallography provides the highest resolution information, which we use to determine the details of binding sites to enable structure-guided drug design.
Synthetic immunology
We utilise protein structures to perform both protein engineering and structure-guided drug design [39].
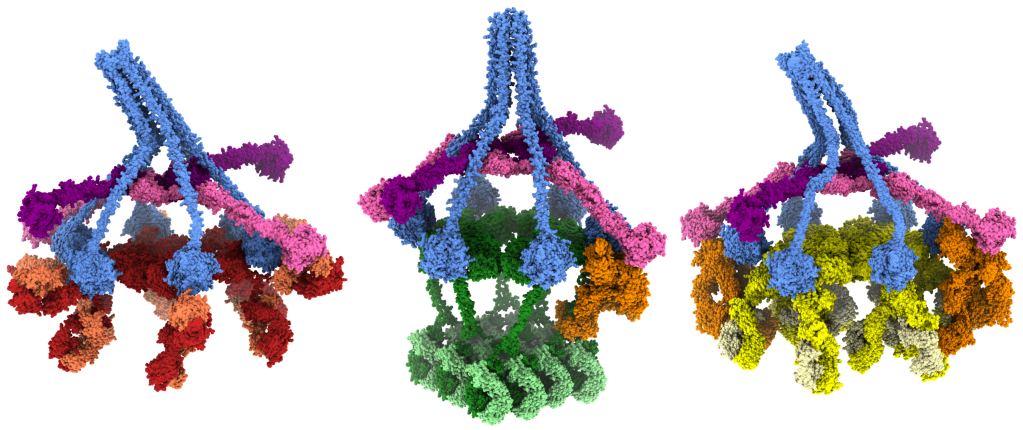
We use DNA nanotechnology to design and synthesise nanoscale scaffolds [24] with defined shapes. We use these scaffolds to template proteins, peptides or small molecules in order to control the geometry of protein binding, which allows us to explore the geometric requirements and constraints of immune system activation. We have used DNA nanotemplates to determine structure-function relationships of antibody-mediated complement activation [41, 44] and intracellular induction of apoptosis [42, 48].
Hear more about the background to some of our DNA nanotechnology research in my TEDx talk.
Methods development for in situ structural biology
We have developed equipment [38], workflows [20, 22] and software [34, 47] to combine the fields of super-resolution light microscopy with cryoEM to achieve high-accuracy localization of tagged proteins within samples prepared for cryoEM [20, 37]. This will allow us to perform structural biology on individual proteins within cells.